Professor Claudio Avignone Rossa FRSB
About
Biography
I obtained my BSc degree in Chemistry (1987), followed by a Licentiate Degree in Biochemical Sciences (1989) and a PhD in Biochemical Sciences (1994) from the University of La Plata (Argentina). I was appointed Associate Professor of Biotechnology at the University of Quilmes (Argentina) in 1994, and Research Fellow at the Microbial Physiology Group at the University of Amsterdam from 1995 to 1999. I joined the University of Surrey in 1999, where I am now a Professor of Systems Microbiology.
Areas of specialism
University roles and responsibilities
- Programme Director, MSc Biotechnology
- Chair, Board of Examiners Level 6
Affiliations and memberships
News
In the media
ResearchResearch interests
Systems microbiology
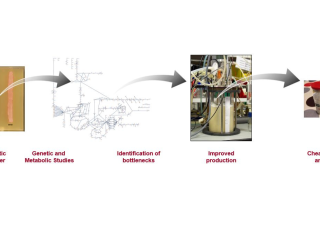
My research interests are in the field of Quantitative Microbial Physiology, Metabolic Modelling and Metabolic Engineering, with the goal of rationally improving the capability of microorganisms for the production of compounds of medical and industrial interest.
- In silico analysis of metabolic networks for the prediction of metabolic capabilities
- Metabolic modelling and quantitative physiology of microorganisms for the production of bioactive molecules
- Metabolic Engineering of microorganisms for the improvement of biosynthetic activities.
The projects combine genomic data, metabolic network modeling, metabolic flux analysis and fermentation technology to design better strategies for antibiotic production, either by the targeted manipulation of specific metabolic pathways or by the modification of the production bioprocess.
Microbial communities
Part of my research is directed to the study of the mechanisms involved in the development and evolution of microbial consortia involved in natural or artificial biological processes. In particular, I am interested in the use of electrogenic microbial communities in bioelectrochemical systems: Microbial fuel cells (MFCs), where micro-organisms in the anodic compartment of a fuel cell produce electricity from organic materials, and Microbial Electrosynthesis Cells (MECs), where an electric current is applied to the system to steer microbial metabolism towards the production of molecules of interest.
Within this area of research, I am interested in the utilisation of microbial communities for the treatment and conversion of agriwaste and wastewater, and the development of clean bioprocesses.
Grants and funding
Current
- Engineering Streptomyces bacteria for the sustainable manufacture of antibiotics. Engineering Biology Mission Award BBSRC (Hoskisson – Strathclyde; Kendrew – GSK; Avignone Rossa – Surrey).
- SonoBio: Microbes that listen: Sono-bio technology for persistent organic pollutants. EPSRC - UKRI Cross Research Council Scheme. (Bussemaker (PI), Sears, Avignone Rossa - Surrey).
- BioRem: Systems Biology, Artificial Intelligence and Advanced Biotechnology Approaches to Improve Soil Bioremediation. HORIZON TMA MSCA (EU) - (University of Burgos - SP; Josef Stefan Institute - SL; Wageningen University of Research - NL; University of Surrey - UK; KEPLER Ingenieria y Ecogestion - SP; LIMNOS Podjetje za Aplikativno Ekologijo - SL)
Recent
- BioElectrochemical LIthium rEcoVEry (BELIEVE). BBSRC.Metabolic modelling and Machine learning for optimization of an industrial enzyme production process. University of Surrey and GSK. PI
- Coupling of photocatalysis and biodegradation for emerging contaminant removal from water effluents and renewable energy generation. Newton Mosharafa / British Council. PI
- GREENER - InteGRated systems for Effective ENvironmEntal Remediation. EU - Horizon 2020. PI in Surrey
- Evaluation of the bioelectrochemical performance of a native microbial community for waste degradation and energy recovery from industrial coffee waste in a low cost microbial fuel cell. Newton Prize. PI.
- Synthetic Biology for Biotechnology and Bioenergy. International Partnering Award BBSRC. PI.
- Metabolic analysis of the solventogenic bacterium Clostridium saccharoperbutylacetonicum. BBSRC iCASE. PI
- Constructing a microbial community for increasing wheat crop yield using system approaches. BBSRC iCASE.Co-I
- Metabolic analysis to characterise and optimize an industrial enzyme production process. BBSRC iCASE. PI
- A bioelectrochemical system for waste degradation and energy recovery from coffee waste. Newton Fund and University of Antioquia, Colombia. PI
Research projects
Engineering Streptomyces bacteria for the sustainable manufacture of antibiotics (current)Engineering Biology Mission Award BBSRC (Hoskisson – Strathclyde; Kendrew – GSK; Avignone Rossa – Surrey).
This project addresses engineering biology for clean growth (primary) and biomedicine. This Engineering Biology Mission Award aims to utilise engineering biology approaches for the development of transferable strategies to engineer antibiotic-producing Streptomyces bacteria able to use a wide range of sustainable feedstocks for production of drug molecules. To deliver this mission we will integrate ‘wet’ and ‘dry’ laboratory data from an authentic industrial Streptomyces strain lineage. We will determine how the strain improvement process has driven strains to adapt to specific media and fermentation conditions and then use this information to build, design and test appropriate engineering strategies for the development and construction of Streptomyces strains to utilise greener carbon sources. This will ultimately enable us to learn how to rationally design strains for greener manufacturing and make this strategy applicable to other industrial Streptomyces strains.
The project will enable antimicrobial production to be cheaper and more sustainable, delivering a less carbon-intensive process for biomanufacturing. It will also be translatable to other products made by Streptomyces bacteria, such as, anti-parasitic, anti-cancer, anti-fungal and immunosuppressant drugs.
SonoBio: Microbes that listen: Sono-bio technology for persistent organic pollutants (current)EPSRC - UKRI Cross Research Council Scheme. (Bussemaker (PI), Sears, Avignone Rossa - Surrey)
VISION: Our vision is to demonstrate a new hybrid technology combining high-frequency ultrasound (sono) and microbes (bio) for the removal of persistent organic pollutants. A new synergy is proposed from simultaneous microbial and ultrasonic action, created through complementary degradation methods and enhanced microbial metabolism in ultrasonic fields.
BACKGROUND: The sono-bio process will be demonstrated on per-and poly-fluorinated alkyl substances (PFAS). PFAS are extremely difficult to destroy, persist in the environment and can be toxic to human and animal health. Environmental bacteria can degrade larger PFAS compounds, albeit very slowly. As yet, complete mineralisation via biological mechanisms i.e. breakdown PFAS to fluoride ions, carbon dioxide and water has not been realised. Sonolysis, via high frequency ultrasound (HFUS, 100-1000 kHz) is one of few technologies demonstrated to fully mineralise PFAS. However, sonolysis is challenged by larger PFAS compounds in mixed matrices. We hypothesise that a combination of ultrasonic and microbial treatments working in synergy, will ultimately deliver sustainable and efficient treatment for PFAS remediation.
AIMS: Sono-bio processing will be demonstrated with microbial processes that benefit from high frequency ultrasonic cavitation, each acting to degrade PFAS via complementary mechanisms. Resource and energy recovery will be achieved by further combination of the process with anode-respiring bacteria in a microbial-electrochemical cell. Novel analytical tools will be used to understand the interaction of ultrasonic cavitation with bacteria.
The objectives of the project include:
- Research interaction of HFUS parameters (e.g. frequency, power, reactor configuration) and different microbial population viabilities using model PFAS.
- Research treatment in mixed matrices using select sono-bio processing configurations.
- Engineer a platform combining sono-bio processing and resource recovery.
- Apply novel analytical tools i) for PFAS analysis in mixed matrices, and ii) to understand cell metabolism in populations versus individual cells.
HORIZON TMA MSCA (EU) - (University of Burgos - SP; Josef Stefan Institute - SL; Wageningen University of Research - NL; University of Surrey - UK; KEPLER Ingenieria y Ecogestion - SP; LIMNOS Podjetje za Aplikativno Ekologijo - SL)
The growing presence of hazardous compounds in the environment such as persistent organic pollutants compromises the health of ecosystems and humans worldwide. The spontaneous ecological recovery of contaminated sites is possible due to the action of biological agents, including plants and microorganisms. The exploitation of the capability of the latter to transform toxic contaminants into harmless end-products can lead to cheap and sustainable bioremediation alternatives. However, the significant knowledge gap on the molecular mechanisms and microbial species responsible for an efficient detoxification of specific pollutants in determined environmental conditions is a burden slowing down the development of efficient microbial assisted bioremediation technologies. BIOREM is an integrated action conformed by experts in microbial systems biology, artificial intelligence tools and environmental sciences that will work together to gain knowledge in the identification of responsible microbial metabolic routes within natural and synthetic consortia for the degradation of target contaminants. The project through inter-sectorial and multidisciplinary training and collaboration will investigate the synergetic effect of different and combined bioremediation strategies, such as bioaugmentation, bioestimulation and microbial-assisted phytoremediation, stablishing links between effective pollutants removal and the responsible microbial pathways. Predictive models for TPHs and PAH remediation will be developed using High-Perfomance Computing (HPC) and Artificial Intelligence to enhance the efficiency of bioremediation strategies by enabling the analysis of vast amounts of environmental data. The integration of the project information (key microbial players and environmental conditions) into mathematical models will allow the establishment of tailored and efficient removal strategies based on the chemical composition and natural microbiome presence in polluted sites.
GREENER is a multi-disciplinary project, involving 15 EU universities, research institutes and SMEs and 5 academic and scientific partners from China, to address the Horizon 2020 topic CE-BIOTEC-04-2018: New biotechnologies for environmental remediation.
GREENER proposes the development of green, sustainable, efficient, and low-cost solutions for soil/sediment and water bioremediation that, by integrating several remediation strategies with innovative bio-electrochemical technologies, will effectively accelerate the remediation time of a range of organic and inorganic pollutants of high concern, while producing end-products of interests, such as bioelectricity and/or harmless metabolites of industrial interest. To achieve such an ambitious goal, organisms with high bioremediation ability will be identified and isolated, the influence of physico-chemical factors on the effectiveness of treatment will be evaluated and proof-of-concept experiments to define optimal integrated solutions at the lab-scale will be performed. Finally, a combination of the most promising technologies will be up-scaled and tested on field. Life cycle analyses will demonstrate the technical and economic feasibility of the solutions suggested.
Increasing chemical pollution seriously compromises the health of ecosystems and humans worldwide. Hazardous compounds, such as polycyclic aromatic hydrocarbons, heavy metals and emerging pollutants contaminate soils/sediments, ground and surface waters. To prevent/minimise the risks associated with the accumulations of these chemicals in the environment it is key to establish low-cost/green methodologies for the treatment and redevelopment of contaminated areas. Several physico-chemical methods have been explored to remove pollutants in the environment, but these are complex, energy consuming or expensive. The exploitation of the capability of bacteria, fungi and phototrophs to transform toxic contaminants into harmless end-products, can lead instead to cheap and sustainable bioremediation alternatives. GREENER proposes the development of innovative, efficient and low-cost hybrid solutions that integrate bioremediation technologies with bio-electrochemical systems (BES). BES, such as microbial fuel cells, break down organic contaminants through the action of electroactive bacteria while generating electrical current. We will investigate the synergetic effect of different bioremediation strategies and demonstrate effective pollutants removal in water and soil/sediments, while generating side products of interest, such as bioelectricity. The type and entity of contamination, along with the specific physico-chemical/microbial characteristics of the environment to be depolluted, will feed into a decision-making toolbox. The latter will allow the establishment of ad hoc integrated solutions, which will take into account effectiveness of biodegradation, costs, environmental risks and social aspects. Fundamental research will be performed at lab-scale, while pilot-tests will be used to proof the scaling-up feasibility for field applications. Environmental benefits and risks, compared to standard remediation approaches, including energy efficiency, will be investigated.
Past project: Coupling of photocatalysis and biodegradation for emerging contaminant removal from water effluents and renewable energy generationBritish Council - Newton Mosharrafa Fund
Current wastewater treatment technologies are efficient at removing most organic contaminants, but consume significant amounts of energy (~1.2 kWh m-3), which makes it an expensive process. Theoretically, the wastewater organic content is sufficient to generate approximately 4 times more energy than is required for wastewater treatment. Thus, it could be possible to minimize energy consumption through capturing part of the energy contained in organic waste streams. A nascent technology to extract energy and value- added chemicals in organic waste streams is the “microbial electrochemical cell” (MXC), a platform technology able to recover energy as electrical current and H2, based on the ability of anode-respiring bacteria (ARB) to oxidize organic matter internally and transfer the resulting electrons to a solid electron acceptor (the anode).
We propose to couple photocatalysis and biodegradation by developing a microbial electrochemical cell (ICPB-MXC) combining visible-light-adsorbing photocatalysts and anode- respiring bacteria to accelerate the degradation of recalcitrant organic compounds while recovering electrical current and hydrogen.
The overarching goal of this collaborative project is to develop an advanced engineered platform combining chemical, visible-light- induced photocatalysis and biodegradation, to accelerate the degradation of recalcitrant organic compounds in wastewater while recovering electrical current and H2.
Newton-Caldas Fund and UK-Colombia Newton Prize 2018
Production of bioelectricity from organic matter has shown great potential as an opportunity to reduce the inefficient disposal and accumulation of organic waste. Despite significant advances in MFCs, further research is required to enhance their performance.
This project will focus on evaluation of bioelectrochemical performance of a native microbial community for waste degradation and energy recovery from industrial coffee waste in a low cost microbial fuel cell. With the design and construction of a low cost microbial fuel cell (MFC) useful in the treatment of agroindustrial coffee waste and the establishment of a native microbial community with high degradation capability for agroindustrial coffee waste. We intend to show great potential to reduce the inefficient disposal and accumulation of organic waste.
The intended result of the project is the development of prototype of an operational microbial fuel cell (MFC) to be used on agro industrial coffee waste. It will be built considering economic and environmental sustainability and its potential application in coffee production farms. We intend to identify a suitable microbial/microorganism strain with high degradation capability of solid-liquid residues and generation of bioelectricity. Additionally, we also intend to define the best operational conditions and performance parameters for a best degradation of these agro industrial residues while generating energy at the same time.
Indicators of esteem
- UKRI - BBSRC Member of BBSRC Pool of Experts
Editor, FEMS Microbiology Reviews
Member of the Scientific Advisory Board, Microbial/Enzymatic Electrochemistry Platform (MEEP)
- Member, Scientific Committee of the Network of Argentine Scientists in the United Kingdom (Red de Científicos Argentinos en el Reino Unido)
Member of the Editorial Board of Applied Microbiology
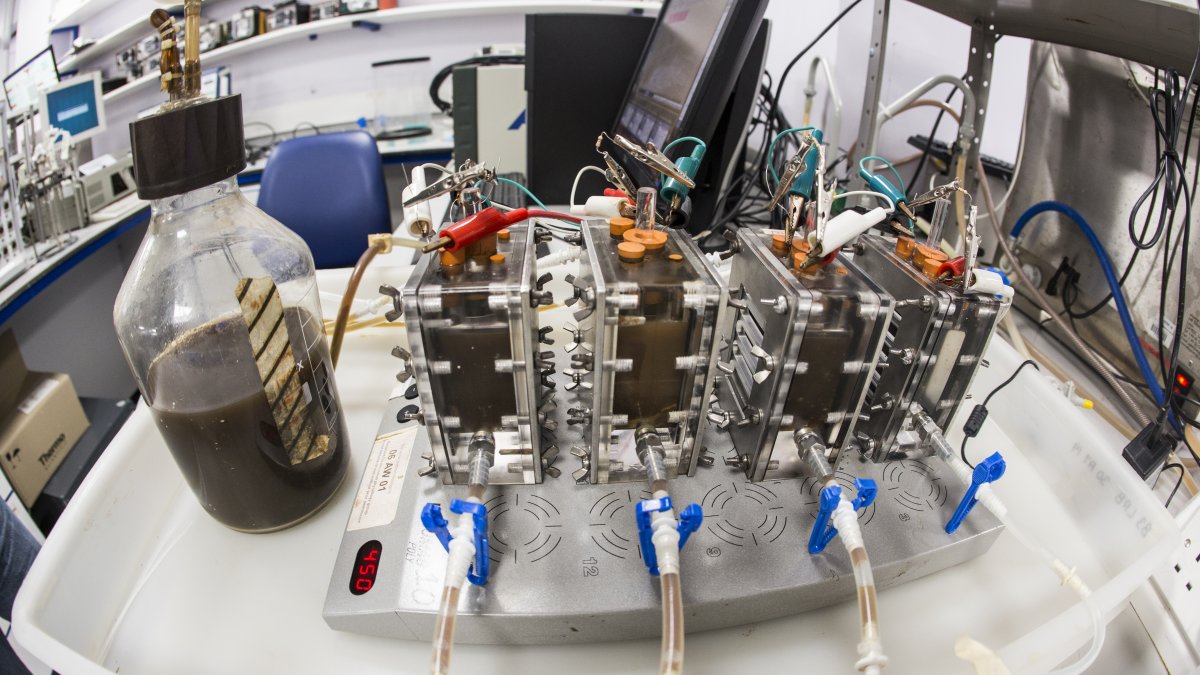
Microbial fuel Cell Cascade fed with wastewater from primary coffee production
 Jaime Gonzalez Gutierrez de la Concha
Jaime Gonzalez Gutierrez de la Concha
Research interests
Systems microbiology
My research interests are in the field of Quantitative Microbial Physiology, Metabolic Modelling and Metabolic Engineering, with the goal of rationally improving the capability of microorganisms for the production of compounds of medical and industrial interest.
- In silico analysis of metabolic networks for the prediction of metabolic capabilities
- Metabolic modelling and quantitative physiology of microorganisms for the production of bioactive molecules
- Metabolic Engineering of microorganisms for the improvement of biosynthetic activities.
The projects combine genomic data, metabolic network modeling, metabolic flux analysis and fermentation technology to design better strategies for antibiotic production, either by the targeted manipulation of specific metabolic pathways or by the modification of the production bioprocess.
Microbial communities
Part of my research is directed to the study of the mechanisms involved in the development and evolution of microbial consortia involved in natural or artificial biological processes. In particular, I am interested in the use of electrogenic microbial communities in bioelectrochemical systems: Microbial fuel cells (MFCs), where micro-organisms in the anodic compartment of a fuel cell produce electricity from organic materials, and Microbial Electrosynthesis Cells (MECs), where an electric current is applied to the system to steer microbial metabolism towards the production of molecules of interest.
Within this area of research, I am interested in the utilisation of microbial communities for the treatment and conversion of agriwaste and wastewater, and the development of clean bioprocesses.
Grants and funding
Current
- Engineering Streptomyces bacteria for the sustainable manufacture of antibiotics. Engineering Biology Mission Award BBSRC (Hoskisson – Strathclyde; Kendrew – GSK; Avignone Rossa – Surrey).
- SonoBio: Microbes that listen: Sono-bio technology for persistent organic pollutants. EPSRC - UKRI Cross Research Council Scheme. (Bussemaker (PI), Sears, Avignone Rossa - Surrey).
- BioRem: Systems Biology, Artificial Intelligence and Advanced Biotechnology Approaches to Improve Soil Bioremediation. HORIZON TMA MSCA (EU) - (University of Burgos - SP; Josef Stefan Institute - SL; Wageningen University of Research - NL; University of Surrey - UK; KEPLER Ingenieria y Ecogestion - SP; LIMNOS Podjetje za Aplikativno Ekologijo - SL)
Recent
- BioElectrochemical LIthium rEcoVEry (BELIEVE). BBSRC.Metabolic modelling and Machine learning for optimization of an industrial enzyme production process. University of Surrey and GSK. PI
- Coupling of photocatalysis and biodegradation for emerging contaminant removal from water effluents and renewable energy generation. Newton Mosharafa / British Council. PI
- GREENER - InteGRated systems for Effective ENvironmEntal Remediation. EU - Horizon 2020. PI in Surrey
- Evaluation of the bioelectrochemical performance of a native microbial community for waste degradation and energy recovery from industrial coffee waste in a low cost microbial fuel cell. Newton Prize. PI.
- Synthetic Biology for Biotechnology and Bioenergy. International Partnering Award BBSRC. PI.
- Metabolic analysis of the solventogenic bacterium Clostridium saccharoperbutylacetonicum. BBSRC iCASE. PI
- Constructing a microbial community for increasing wheat crop yield using system approaches. BBSRC iCASE.Co-I
- Metabolic analysis to characterise and optimize an industrial enzyme production process. BBSRC iCASE. PI
- A bioelectrochemical system for waste degradation and energy recovery from coffee waste. Newton Fund and University of Antioquia, Colombia. PI
Research projects
Engineering Biology Mission Award BBSRC (Hoskisson – Strathclyde; Kendrew – GSK; Avignone Rossa – Surrey).
This project addresses engineering biology for clean growth (primary) and biomedicine. This Engineering Biology Mission Award aims to utilise engineering biology approaches for the development of transferable strategies to engineer antibiotic-producing Streptomyces bacteria able to use a wide range of sustainable feedstocks for production of drug molecules. To deliver this mission we will integrate ‘wet’ and ‘dry’ laboratory data from an authentic industrial Streptomyces strain lineage. We will determine how the strain improvement process has driven strains to adapt to specific media and fermentation conditions and then use this information to build, design and test appropriate engineering strategies for the development and construction of Streptomyces strains to utilise greener carbon sources. This will ultimately enable us to learn how to rationally design strains for greener manufacturing and make this strategy applicable to other industrial Streptomyces strains.
The project will enable antimicrobial production to be cheaper and more sustainable, delivering a less carbon-intensive process for biomanufacturing. It will also be translatable to other products made by Streptomyces bacteria, such as, anti-parasitic, anti-cancer, anti-fungal and immunosuppressant drugs.
EPSRC - UKRI Cross Research Council Scheme. (Bussemaker (PI), Sears, Avignone Rossa - Surrey)
VISION: Our vision is to demonstrate a new hybrid technology combining high-frequency ultrasound (sono) and microbes (bio) for the removal of persistent organic pollutants. A new synergy is proposed from simultaneous microbial and ultrasonic action, created through complementary degradation methods and enhanced microbial metabolism in ultrasonic fields.
BACKGROUND: The sono-bio process will be demonstrated on per-and poly-fluorinated alkyl substances (PFAS). PFAS are extremely difficult to destroy, persist in the environment and can be toxic to human and animal health. Environmental bacteria can degrade larger PFAS compounds, albeit very slowly. As yet, complete mineralisation via biological mechanisms i.e. breakdown PFAS to fluoride ions, carbon dioxide and water has not been realised. Sonolysis, via high frequency ultrasound (HFUS, 100-1000 kHz) is one of few technologies demonstrated to fully mineralise PFAS. However, sonolysis is challenged by larger PFAS compounds in mixed matrices. We hypothesise that a combination of ultrasonic and microbial treatments working in synergy, will ultimately deliver sustainable and efficient treatment for PFAS remediation.
AIMS: Sono-bio processing will be demonstrated with microbial processes that benefit from high frequency ultrasonic cavitation, each acting to degrade PFAS via complementary mechanisms. Resource and energy recovery will be achieved by further combination of the process with anode-respiring bacteria in a microbial-electrochemical cell. Novel analytical tools will be used to understand the interaction of ultrasonic cavitation with bacteria.
The objectives of the project include:
- Research interaction of HFUS parameters (e.g. frequency, power, reactor configuration) and different microbial population viabilities using model PFAS.
- Research treatment in mixed matrices using select sono-bio processing configurations.
- Engineer a platform combining sono-bio processing and resource recovery.
- Apply novel analytical tools i) for PFAS analysis in mixed matrices, and ii) to understand cell metabolism in populations versus individual cells.
HORIZON TMA MSCA (EU) - (University of Burgos - SP; Josef Stefan Institute - SL; Wageningen University of Research - NL; University of Surrey - UK; KEPLER Ingenieria y Ecogestion - SP; LIMNOS Podjetje za Aplikativno Ekologijo - SL)
The growing presence of hazardous compounds in the environment such as persistent organic pollutants compromises the health of ecosystems and humans worldwide. The spontaneous ecological recovery of contaminated sites is possible due to the action of biological agents, including plants and microorganisms. The exploitation of the capability of the latter to transform toxic contaminants into harmless end-products can lead to cheap and sustainable bioremediation alternatives. However, the significant knowledge gap on the molecular mechanisms and microbial species responsible for an efficient detoxification of specific pollutants in determined environmental conditions is a burden slowing down the development of efficient microbial assisted bioremediation technologies. BIOREM is an integrated action conformed by experts in microbial systems biology, artificial intelligence tools and environmental sciences that will work together to gain knowledge in the identification of responsible microbial metabolic routes within natural and synthetic consortia for the degradation of target contaminants. The project through inter-sectorial and multidisciplinary training and collaboration will investigate the synergetic effect of different and combined bioremediation strategies, such as bioaugmentation, bioestimulation and microbial-assisted phytoremediation, stablishing links between effective pollutants removal and the responsible microbial pathways. Predictive models for TPHs and PAH remediation will be developed using High-Perfomance Computing (HPC) and Artificial Intelligence to enhance the efficiency of bioremediation strategies by enabling the analysis of vast amounts of environmental data. The integration of the project information (key microbial players and environmental conditions) into mathematical models will allow the establishment of tailored and efficient removal strategies based on the chemical composition and natural microbiome presence in polluted sites.
GREENER is a multi-disciplinary project, involving 15 EU universities, research institutes and SMEs and 5 academic and scientific partners from China, to address the Horizon 2020 topic CE-BIOTEC-04-2018: New biotechnologies for environmental remediation.
GREENER proposes the development of green, sustainable, efficient, and low-cost solutions for soil/sediment and water bioremediation that, by integrating several remediation strategies with innovative bio-electrochemical technologies, will effectively accelerate the remediation time of a range of organic and inorganic pollutants of high concern, while producing end-products of interests, such as bioelectricity and/or harmless metabolites of industrial interest. To achieve such an ambitious goal, organisms with high bioremediation ability will be identified and isolated, the influence of physico-chemical factors on the effectiveness of treatment will be evaluated and proof-of-concept experiments to define optimal integrated solutions at the lab-scale will be performed. Finally, a combination of the most promising technologies will be up-scaled and tested on field. Life cycle analyses will demonstrate the technical and economic feasibility of the solutions suggested.
Increasing chemical pollution seriously compromises the health of ecosystems and humans worldwide. Hazardous compounds, such as polycyclic aromatic hydrocarbons, heavy metals and emerging pollutants contaminate soils/sediments, ground and surface waters. To prevent/minimise the risks associated with the accumulations of these chemicals in the environment it is key to establish low-cost/green methodologies for the treatment and redevelopment of contaminated areas. Several physico-chemical methods have been explored to remove pollutants in the environment, but these are complex, energy consuming or expensive. The exploitation of the capability of bacteria, fungi and phototrophs to transform toxic contaminants into harmless end-products, can lead instead to cheap and sustainable bioremediation alternatives. GREENER proposes the development of innovative, efficient and low-cost hybrid solutions that integrate bioremediation technologies with bio-electrochemical systems (BES). BES, such as microbial fuel cells, break down organic contaminants through the action of electroactive bacteria while generating electrical current. We will investigate the synergetic effect of different bioremediation strategies and demonstrate effective pollutants removal in water and soil/sediments, while generating side products of interest, such as bioelectricity. The type and entity of contamination, along with the specific physico-chemical/microbial characteristics of the environment to be depolluted, will feed into a decision-making toolbox. The latter will allow the establishment of ad hoc integrated solutions, which will take into account effectiveness of biodegradation, costs, environmental risks and social aspects. Fundamental research will be performed at lab-scale, while pilot-tests will be used to proof the scaling-up feasibility for field applications. Environmental benefits and risks, compared to standard remediation approaches, including energy efficiency, will be investigated.
British Council - Newton Mosharrafa Fund
Current wastewater treatment technologies are efficient at removing most organic contaminants, but consume significant amounts of energy (~1.2 kWh m-3), which makes it an expensive process. Theoretically, the wastewater organic content is sufficient to generate approximately 4 times more energy than is required for wastewater treatment. Thus, it could be possible to minimize energy consumption through capturing part of the energy contained in organic waste streams. A nascent technology to extract energy and value- added chemicals in organic waste streams is the “microbial electrochemical cell” (MXC), a platform technology able to recover energy as electrical current and H2, based on the ability of anode-respiring bacteria (ARB) to oxidize organic matter internally and transfer the resulting electrons to a solid electron acceptor (the anode).
We propose to couple photocatalysis and biodegradation by developing a microbial electrochemical cell (ICPB-MXC) combining visible-light-adsorbing photocatalysts and anode- respiring bacteria to accelerate the degradation of recalcitrant organic compounds while recovering electrical current and hydrogen.
The overarching goal of this collaborative project is to develop an advanced engineered platform combining chemical, visible-light- induced photocatalysis and biodegradation, to accelerate the degradation of recalcitrant organic compounds in wastewater while recovering electrical current and H2.
Newton-Caldas Fund and UK-Colombia Newton Prize 2018
Production of bioelectricity from organic matter has shown great potential as an opportunity to reduce the inefficient disposal and accumulation of organic waste. Despite significant advances in MFCs, further research is required to enhance their performance.
This project will focus on evaluation of bioelectrochemical performance of a native microbial community for waste degradation and energy recovery from industrial coffee waste in a low cost microbial fuel cell. With the design and construction of a low cost microbial fuel cell (MFC) useful in the treatment of agroindustrial coffee waste and the establishment of a native microbial community with high degradation capability for agroindustrial coffee waste. We intend to show great potential to reduce the inefficient disposal and accumulation of organic waste.
The intended result of the project is the development of prototype of an operational microbial fuel cell (MFC) to be used on agro industrial coffee waste. It will be built considering economic and environmental sustainability and its potential application in coffee production farms. We intend to identify a suitable microbial/microorganism strain with high degradation capability of solid-liquid residues and generation of bioelectricity. Additionally, we also intend to define the best operational conditions and performance parameters for a best degradation of these agro industrial residues while generating energy at the same time.
Indicators of esteem
- UKRI - BBSRC Member of BBSRC Pool of Experts
Editor, FEMS Microbiology Reviews
Member of the Scientific Advisory Board, Microbial/Enzymatic Electrochemistry Platform (MEEP)
- Member, Scientific Committee of the Network of Argentine Scientists in the United Kingdom (Red de Científicos Argentinos en el Reino Unido)
Member of the Editorial Board of Applied Microbiology
Microbial fuel Cell Cascade fed with wastewater from primary coffee production
 Jaime Gonzalez Gutierrez de la Concha
Jaime Gonzalez Gutierrez de la ConchaTeaching
I am Programme Director and Lecturer of the MSc Biotechnology course, and Module Organizer and Lecturer of the module Biomedical Microbial Products (BMS3060), and lecturer in the modules Microbial Communities and Interactions (BMS3115) and Introduction to the Microbial World (BMS1035)
Sustainable development goals
My research interests are related to the following:





Publications
Publications since 2020
- Martinez Martin et al (2025) Exsolution of Pt Nanoparticles from Mixed Zr/Gd‐CeO2 Oxides for Microbial Fuel Cell‐Based Biosensors. Small Science, 2400619
- Salvian et al (2025) Cost-effective and stable biosensing of biochemical oxygen demand in wastewater treatment: Exsolved rhodium-titanate perovskite catalyst in microbial fuel cell-based biosensors. Journal of Environmental Chemical Engineering 13 (2), 115692
- Nastro et al (2025) Bio-electrosynthesis of polyhydroxybutyrate and surfactants in microbial fuel cells: a preliminary study. Frontiers in Microbiology 16, 1372302
- Wang et al (2025) Multi-faceted effects and mechanisms of granular activated carbon to enhance anaerobic ammonium oxidation (anammox) for nitrogen removal from wastewater. Bioresource technology 418, 132001
- Salvian et al; (2025) 3D‐Printed Macroporous Resin Anode in Microbial Fuel Cell‐Based Biosensors for Efficient Biodegradable Organic Carbon Monitoring. Advanced Sensor Research, e00072
- Farkas et al (2024) Assessing the impact of soil microbial fuel cells on atrazine removal in soil. Journal of Hazardous Materials 478, 135473
- Salvian et al (2024) Impact of air-cathodes on operational stability of single-chamber microbial fuel cell biosensors for wastewater monitoring. Energies 17(14), 3574
- Salvian et al (2024) Resilience of anodic biofilm in microbial fuel cell biosensor for BOD monitoring of urban wastewater. npj Clean Water 7(1), 53.
- Perez-de-Mora et al (2024). Biostimulation of sulfate reduction for in-situ metal (loid) precipitation at an industrial site in Flanders, Belgium. Science of The Total Environment 929, 172298
- Rafieenia et al (2024) , Enhanced electricity generation in solar-driven photo-bioelectrochemical cells equipped with Co3 (PO4) 2/Mg (OH) 2 photoanode. Water 16(12), 1683
- Garcia-Mogollon et al (2023) C. saccharoperbutylacetonicum n1-4 electroactivity and co2 fixation under different electrochemical conditions. Rasayan J. Chem. 16 (4), 2057-2063
- Nandy et al (2023) Influence of carbon-based cathodes on biofilm composition and electrochemical performance in soil microbial fuel cells. Environmental Science and Ecotechnology 16, 100276
- Oluchukwu et al (2023) Yam peels as potential feed ingredients in poultry diets: Nutritional composition, physical properties and prospects in circular economy. Zenodo https://doi.org/10.5281/zenodo.8051390
- Nastro et al (2023) Inorganic Carbon Assimilation and Electrosynthesis of Platform Chemicals in Bioelectrochemical Systems (BESs) Inoculated with Clostridium saccharoperbutylacetonicum N1-H4. Microorganisms 11, 735
- Kordek-Khali et al (2023) Nanocomposite use in MFCs: a state of the art review. Sustainable Energy & Fuels 7 (24), 5608-5624
- Rafieenia et al (2022) The degradation of glyphosate is enhanced in a microbial fuel cell: Electrochemical performance, degradation efficiency, and analysis of the anodic microbial community. Sustainable Energy Technologies and Assessments 54, 102805
- Rafieenia et al. (2022) Integration of microbial electrochemical systems and photocatalysis for sustainable treatment of organic recalcitrant wastewaters. Science of The Total Environment 824, 153923
- Agudelo Escobar et al (2022). A Bioelectrochemical system for waste degradation and energy recovery from industrial coffee wastewater. Front. Chem. Eng. 4:814987. doi: 10.3389/fceng.2022.814987.
- Rafieenia et al (2022). Integration of microbial electrochemical systems and photocatalysis for sustainable treatment of organic recalcitrant wastewaters: Main mechanisms, recent advances, and present prospects. Science of the Total Environment 824: 153923
- González et al (2021) Loss of a pyoverdine secondary receptor in Pseudomonas aeruginosa results in a fitter strain suitable for population invasion. The ISME Journal. https://doi.org/10.1038/s41396-020-00853-2
- Yusuf et al (2020). Valorisation of banana peels by hydrothermal carbonisation: Potential use of the hydrochar and liquid by-product for water purification and energy conversion. Biores Technology Rep. https://doi.org/10.1016/j.biteb.2020.100582.
- Endreny et al (2020) Generating electricity with urban green infrastructure microbial fuel cells. J Cleaner Prod 263,121337. https://doi.org/10.1016/j.jclepro.2020.121337.