Dr Aurore Poirier
About
Biography
Aurore is a Research Fellow B in Molecular Microbiology and a Science Communication officer, as part of the One Health EJP consortium. She joined the University of Surrey in 2018, in Professor La Ragione’s research group and has been working since then on the development of rapid molecular diagnostics for infectious diseases.
Aurore successively worked as a Postdoctoral Research Fellow in Molecular Microbiology on a BBSRC funded project, to develop a rapid diagnostics platform for the detection of bacterial and viral pathogens in broiler chickens in the Philippines, in collaboration with Brunel University London, Lancaster University and 3 Universities of The Philippines; and on an Innovate UK project for the development of a New Smart Diagnostic for Infection with Prof Roberto La Ragione and Prof Johnjoe McFadden, in collaboration with Molecular Warehouse and Shanghai University.
Before joining the University of Surrey, Aurore was, for 2 years, a Research Associate working on the development of a bio-medical device to treat sepsis, with Pr Clare Selden in the Institute for Liver and Digestive Health at University College London (UCL). She proved the efficiency of treatment with the developed prototype on human plasma infected with Gram-negative bacteria.
Aurore was awarded a PhD in Microbiology and Parasitology in 2015, after completing an MSc in Biology of Plants and Micro-organisms in 2012 both at the University of Montpellier, France. She conducted her PhD studies in the Host-Pathogen-Environment Interactions laboratory in Montpellier, under the supervision of Dr Delphine Destoumieux-Garzon and Dr Guillaume Charriere. The project involved the study of the interactions between Vibrio pathogenic for oysters and two different phagocytes: oyster immune cells (haemocytes) and amoebae isolated from the field.
Areas of specialism
University roles and responsibilities
- Research fellow B in Molecular Microbiology
- Science Communication Officer
My qualifications
Previous roles
Business, industry and community links
Publications
Although antimicrobial histones have been isolated from multiple metazoan species, their role in host defense has long remained unanswered. We found here that the hemocytes of the oyster Crassostrea gigas release antimicrobial H1-like and H5-like histones in response to tissue damage and infection. These antimicrobial histones were shown to be associated with extracellular DNA networks released by hemocytes, the circulating immune cells of invertebrates, in response to immune challenge. The hemocyte-released DNA was found to surround and entangle vibrios. This defense mechanism is reminiscent of the neutrophil extracellular traps (ETs) recently described in vertebrates. Importantly, oyster ETs were evidenced in vivo in hemocyte-infiltrated interstitial tissues surrounding wounds, whereas they were absent from tissues of unchallenged oysters. Consistently, antimicrobial histones were found to accumulate in oyster tissues following injury or infection with vibrios. Finally, oyster ET formation was highly dependent on the production of reactive oxygen species by hemocytes. This shows that ET formation relies on common cellular and molecular mechanisms from vertebrates to invertebrates. Altogether, our data reveal that ET formation is a defense mechanism triggered by infection and tissue damage, which is shared by relatively distant species suggesting either evolutionary conservation or convergent evolution within Bilateria.
KEYWORDS:
Oysters are sessile filter feeders that live in close association with abundant and diverse communities of microorganisms that form the oyster microbiota. In such an association, cellular and molecular mechanisms have evolved to maintain oyster homeostasis upon stressful conditions including infection and changing environments. We give here cellular and molecular insights into the Crassostrea gigas antimicrobial defense system with focus on antimicrobial peptides and proteins (AMPs). This review highlights the central role of the hemocytes in the modulation and control of oyster antimicrobial response. As vehicles for AMPs and other antimicrobial effectors, including reactive oxygen species (ROS), and together with epithelia, hemocytes provide the oyster with local defense reactions instead of systemic humoral ones. These reactions are largely based on phagocytosis but also, as recently described, on the extracellular release of antimicrobial histones (ETosis) which is triggered by ROS. Thus, ROS can signal danger and activate cellular responses in the oyster. From the current literature, AMP production/release could serve similar functions. We provide also new lights on the oyster genetic background that underlies a great diversity of AMP sequences but also an extraordinary individual polymorphism of AMP gene expression. We discuss here how this polymorphism could generate new immune functions, new pathogen resistances or support individual adaptation to environmental stresses.
Recent studies revealed that several vibrio species have evolved the capacity to survive inside host cells. However, it is still often ignored if intracellular stages are required for pathogenicity. Virulence of Vibrio tasmaniensis LGP32, a strain pathogenic for Crassostrea gigas oysters, depends on entry into hemocytes, the oyster immune cells. We investigated here the mechanisms of LGP32 intracellular survival and their consequences on the host-pathogen interaction. Entry and survival inside hemocytes were required for LGP32-driven cytolysis of hemocytes, both in vivo and in vitro. LGP32 intracellular stages showed a profound boost in metabolic activity and a major transcription of antioxidant and copper detoxification genes, as revealed by RNA sequencing. LGP32 isogenic mutants showed that resistance to oxidative stress and copper efflux are two main functions required for vibrio intracellular stages and cytotoxicity to hemocytes. Copper efflux was also essential for host colonization and virulence in vivo. Altogether, our results identify copper resistance as a major mechanism to resist killing by phagocytes, induce cytolysis of immune cells and colonize oysters. Selection of such resistance traits could arise from vibrio interactions with copper-rich environmental niches including marine invertebrates, which favour the emergence of pathogenic vibrios resistant to intraphagosomal killing across animal species.
Delivery of cell therapies often requires the ability to hold products in readiness whilst logistical, regulatory and potency considerations are dealt with and recorded. This requires reversibly stopping biological time, a process which is often achieved by cryopreservation. However, cryopreservation itself poses many biological and biophysical challenges to living cells that need to be understood in order to apply the low temperature technologies to their best advantage. This review sets out the history of applied cryopreservation, our current understanding of the various processes involved in storage at cryogenic temperatures, and challenges for robust and reliable uses of cryopreservation within the cell therapy arena.
Vibrios are ubiquitous in marine environments and opportunistically colonize a broad range of hosts. Strains of Vibrio tasmaniensis present in oyster farms can thrive in oysters during juvenile mortality events and behave as facultative intracellular pathogen of oyster haemocytes. Herein, we wondered whether V. tasmaniensis LGP32 resistance to phagocytosis is specific to oyster immune cells or contributes to resistance to other phagocytes, like marine amoebae. To address this question, we developed an integrative study, from the first description of amoeba diversity in oyster farms to the characterization of LGP32 interactions with amoebae. An isolate of the Vannella genus, Vannella sp. AP1411, which was collected from oyster farms, is ubiquitous, and belongs to one clade of Vannella that could be found associated with Vibrionaceae. LGP32 was shown to be resistant to grazing by Vannella sp. AP1411 and this phenotype depends on some previously identified virulence factors: secreted metalloprotease Vsm and copper efflux p‐ATPase CopA, which act at different steps during amoeba–vibrio interactions, whereas some other virulence factors were not involved. Altogether, our work indicates that some virulence factors can be involved in multi‐host interactions of V. tasmaniensis ranging from protozoans to metazoans, potentially favouring their opportunistic behaviour.
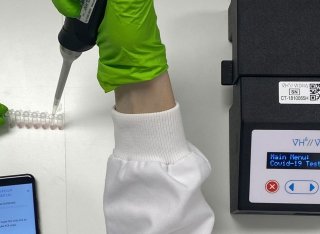
Until vaccines and effective therapeutics become available, the practical solution to transit safely out of the current coronavirus disease 19 (CoVID-19) lockdown may include the implementation of an effective testing, tracing and tracking system. However, this requires a reliable and clinically validated diagnostic platform for the sensitive and specific identification of SARS-CoV-2. Here, we report on the development of a de novo, high-resolution and comparative genomics guided reverse-transcribed loop-mediated isothermal amplification (LAMP) assay. To further enhance the assay performance and to remove any subjectivity associated with operator interpretation of results, we engineered a novel hand-held smart diagnostic device. The robust diagnostic device was further furnished with automated image acquisition and processing algorithms and the collated data was processed through artificial intelligence (AI) pipelines to further reduce the assay run time and the subjectivity of the colorimetric LAMP detection. This advanced AI algorithm-implemented LAMP (ai-LAMP) assay, targeting the RNA-dependent RNA polymerase gene, showed high analytical sensitivity and specificity for SARS-CoV-2. A total of ~200 coronavirus disease (CoVID-19)-suspected NHS patient samples were tested using the platform and it was shown to be reliable, highly specific and significantly more sensitive than the current gold standard qRT-PCR. Therefore, this system could provide an efficient and cost-effective platform to detect SARS-CoV-2 in resource-limited laboratories.
An artificial intelligence-assisted low-cost portable device for the rapid detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is presented here. This standalone temperature-controlled device houses tubes designed for conducting reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays. Moreover, the device utilises tubes illuminated by LEDs, an in-built camera, and a small onboard computer with automated image acquisition and processing algorithms. This intelligent device significantly reduces the normal assay run time and removes the subjectivity associated with operator interpretation of colourimetric RT-LAMP results. To further improve this device’s usability, a mobile app has been integrated into the system to control the LAMP assay environment and to visually display the assay results by connecting the device to a smartphone via Bluetooth. This study was undertaken using ~5000 images produced from the ~200 LAMP amplification assays using the prototype device. Synthetic RNA and a small panel of positive and negative SARS-CoV-2 patient samples were assayed for this study. State-of-the-art image processing and artificial intelligence algorithms were applied to these images to analyse them and to select the most efficient algorithm. The template matching algorithm for image extraction and MobileNet CNN architecture for classification results provided 98.0% accuracy with an average run time of 20 min to confirm the endpoint result. Two working points were chosen based on the best compromise between sensitivity and specificity. The high sensitivity point has a sensitivity value of 99.12% and specificity value of 70.8%, while at the high specificity point, the sensitivity is 96.05% and specificity 93.59%. Furthermore, this device provides an efficient and cost-effective platform for non-health professionals to detect not only SARS-CoV-2 but also other pathogens in resource-limited laboratories, factories, airports, schools, universities, and homes.
Klebsiella pneumoniae is an important pathogenic bacterium commonly associated with human healthcare and community-acquired infections. In recent years, K. pneumoniae has become a significant threat to global public and veterinary health, because of its high rates of antimicrobial resistance (AMR). Early diagnosis of K. pneumoniae infection and detection of any associated AMR would help to accelerate directed therapy and reduce the risk of the emergence of multidrug-resistant isolates. In this study, we identified three target genes (yhaI, epsL, and xcpW) common to K. pneumoniae isolates from both China and Europe and designed loop-mediated isothermal amplification (LAMP) assays for the detection of K. pneumoniae in clinical samples. We also designed LAMP assays for the detection of five AMR genes commonly associated with K. pneumoniae. The LAMP assays were validated on a total of 319 type reference strains and clinical isolates of diverse genetic backgrounds, in addition to 40 clinical human sputum samples, and were shown to be reliable, highly specific, and sensitive. For the K. pneumoniae-specific LAMP assay, the calculated sensitivity, specificity, and positive and negative predictive values (comparison with culture and matrix-assisted laser desorption/ionization-time of flight mass spectrometry) were all 100% on clinical isolates and, respectively, of 100%, 91%, and 90%, and 100% when tested on clinical sputum samples, while being significantly faster than the reference methods. For the bla KPC and other carbapenemases' LAMP assays, the concordance between the LAMP results and the references methods (susceptibility tests) was 100%, on both pure cultures (n = 125) and clinical samples (n = 18). In conclusion, we developed highly sensitive and specific LAMP assays for the clinical identification of K. pneumoniae and detection of carbapenem resistance.
Loop-mediated isothermal amplification (LAMP) is a rapid alternative to PCR, in which the reaction occurs at one temperature and uses a polymerase with high displacement activity, e.g. Bacillus stearothermophilus DNA polymerase I (Bst) or homologues. Since the discovery of LAMP in 2000, several applications have been developed to employ this technique in the rapid detection of nucleic acid targets and enhance its performance. Improvements to the LAMP technique and a variety of innovative detection methods have led to its application for a wide range of targets in medical and veterinary microbiology, particularly in resource-poor settings. The key advantages of LAMP-based diagnostics include the ability to rapidly detect target nucleic acid sequences within 30 min and its ease of use, facilitating its application in field, bedside, pen-side, point-of-care and point-of-need diagnostic settings. LAMP can be a valuable tool to aid in the detection and management of disease outbreaks.
Rapid, reliable, sensitive, portable, and accurate diagnostics are required to control disease outbreaks such as COVID-19 that pose an immense burden on human health and the global economy. Here we developed a loop-mediated isothermal amplification (LAMP)-based electrochemical test for the detection of SARS-CoV-2 that causes COVID-19. The test is based on the oxidation-reduction reaction between pyrophosphates (generated from positive LAMP reaction) and molybdate that is detected by cyclic voltammetry using inexpensive and disposable carbon screen printed electrodes. Our test showed higher sensitivity (detecting as low as 5.29 RNA copies/μL) compared to the conventional fluorescent reverse transcriptase (RT)-LAMP. We validated our tests using human serum and saliva spiked with SARS-CoV-2 RNA and clinical (saliva and nasal-pharyngeal) swab samples demonstrating 100% specificity and 93.33% sensitivity. Our assay provides a rapid, specific, and sensitive test with an electrochemical readout in less than 45 min that could be adapted for point-of-care settings.